DEC OMERO Server: Difference between revisions
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
The collection scanned images are currently available as a test site with only a few sample images loaded. There is no unregistered user access available to the server. All users must apply for an account and agree to abide by our access and reuse conditions before being issued a user account. Individual user accounts currently only have access to specific folders. Access to other collection folders can be managed through user rights management. | The collection scanned images are currently available as a test site with only a few sample images loaded. There is no unregistered user access available to the server. All users must apply for an account and agree to abide by our access and reuse conditions before being issued a user account. Individual user accounts currently only have access to specific folders. Access to other collection folders can be managed through user rights management. | ||
2017 - [http://blog.openmicroscopy.org/future-plans/community/2016/12/21/omero-5-3/ OMERO 5.3 upcoming release] - [http://glencoesoftware.com/2016-08-30-glencoe-software-zeiss-partner-open-source-file-reader-whole-slide.html JPGXR announcement] | |||
:[[Help|'''User Help''']] - [[Help#OMERO_Image_Server_Log_In|Image Server Log in]] | [[Help#DEC_Log_In|DEC Log in]] | :[[Help|'''User Help''']] - [[Help#OMERO_Image_Server_Log_In|Image Server Log in]] | [[Help#DEC_Log_In|DEC Log in]] | ||
Revision as of 13:58, 29 January 2017
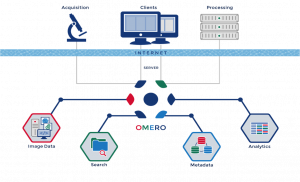
About the Collection Server
| DEC OMERO Server | ||||||||||||||||
| ||||||||||||||||
- Draft page - notice removed when completed. No unregistered user can edit this site or the linked image database. MediaWiki Help page
The collection scanned images are currently available as a test site with only a few sample images loaded. There is no unregistered user access available to the server. All users must apply for an account and agree to abide by our access and reuse conditions before being issued a user account. Individual user accounts currently only have access to specific folders. Access to other collection folders can be managed through user rights management.
2017 - OMERO 5.3 upcoming release - JPGXR announcement
For server issues, access problems or account requests, please contact Dr Mark Hill.
| DEC OMERO Server | ||||||||||||||||
| ||||||||||||||||
Collections have been organised into folders named after their location city, as shown below.
- Amsterdam
- Barcelona
- Berlin
- Bochum
- Göttingen
- Kyoto
- Madrid
- Newcastle
- Sydney
- Washington
OMERO Image Server Log In
Only registered users can log in and edit the OMERO website.
|
This animation shows the described process.
<html5media width="500" height="350">File:OMERO logging in.mp4</html5media> |
Test Image Link
Note - You will need to login to access the original image through the links, images appearing on this current page are only screenshots.
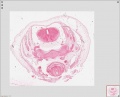
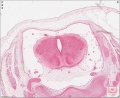
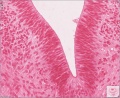
Sydney folder -> 2010-08-29-1 -> Stage 22 Human Embryo (Mark Hill-001.svs) dimensions 32538 x 28770
Image Formats
The original decision was that scanned slides would be saved in JPGXR format as these are substantially smaller than the uncompressed format. The difficulty is that OMERO server does not support this format so all images will now be stored in an uncompressed format that is compatible with this image server software.
Apply for DEC Account
For user account requests, please contact Dr Mark Hill.
Draft Terms of Use
(Could consortium members please comment on these draft terms before I prepare a document that will be send to each applicant for a user account).
By using the DEC OMERO Server, you agree to the following:
- You agree to the specific conditions set by each collections terms of image reuse, publication and copyright.
- Reuse of images from multiple collections requires approval from each collection.
- You use this service at your own risk, and we provide NO GUARANTEE OR WARRANTY WHATSOEVER regarding uptime, security, data provenance or access, or any other service.
- You should not use the DEC OMERO Server as a repository for any important data, or data that must be held in any secure or specific way.
- The responsibility for any data loaded onto the DEC OMERO Server is entirely your own.
- We reserve the right to remove any images, data, accounts or other data or structure without notice or request.
- We welcome comments or ideas from your use of and experience with the DEC OMERO Server.
About OMERO
OMERO is client-server software for visualization, management and analysis of biological microscope images. © 2000-2016 University of Dundee & Open Microscopy Environment. Creative Commons Attribution 4.0 International License OME source code is available under the GNU General public license or through commercial license from Glencoe Software.
Version - OMERO.web 5.0.8-ice35.
Importing Data
Images are added to the database by the process called "Importing Data" using the software OMERO.insight Version 5
Help - Importing Data with OMERO.insight Version 5
References
Melissa Linkert, Curtis T. Rueden, Chris Allan, Jean-Marie Burel, Will Moore, Andrew Patterson, Brian Loranger, Josh Moore, Carlos Neves, Donald MacDonald, Aleksandra Tarkowska, Caitlin Sticco, Emma Hill, Mike Rossner, Kevin W. Eliceiri, and Jason R. Swedlow (2010) Metadata matters: access to image data in the real world. The Journal of Cell Biology, Vol. 189no. 5777-782 doi: 10.1083/jcb.201004104 PMID 20513764
Chris Allan, Jean-Marie Burel, Josh Moore, Colin Blackburn, Melissa Linkert, Scott Loynton, Donald MacDonald, William J Moore, Carlos Neves, Andrew Patterson, Michael Porter, Aleksandra Tarkowska, Brian Loranger, Jerome Avondo, Ingvar Lagerstedt, Luca Lianas, Simone Leo, Katherine Hands, Ron T Hay, Ardan Patwardhan, Christoph Best, Gerard J Kleywegt, Gianluigi Zanetti & Jason R Swedlow (2012) OMERO: flexible, model-driven data management for experimental biology. Nature Methods 9, 245–253. Published: 28 February 2012 PMID 22373911
Links
- The Open Microscopy Environment
- OME Help
- Using OMERO.web
- Training Course Manual
- Importing Data with OMERO.insight Version 5
- JCB DataViewer
Main Page | Embryo Collections | Slide Scanning | Image Server | News | Links | Test page | Site Map